Pre-meeting Events: Development and Cell Biology, MaizeMine, Gramene, MaizeGDB, and Danforth Center workshops!
This year, six pre-conference workshops are being offered prior to the start of the meeting on Thursday, March 16th! Registration is required for the Maize Development Genetics and the Danforth Center tour workshops.
2023 Schedule:
| Thursday, March 16, 2023 |
Contact: |
Location: |
| 9:00 AM - 4:00 PM | Development and Cell Biology Workshop.
Join us for a workshop dedicated to discussing the latest research in Maize Development and Cell Biology. Graduate students, postdocs and junior faculty will present their results in a short talk format, with a small, friendly audience. Plenty of time will be available after each talk for animated discussion. If you would like to present your work, or if you have any questions, please email Paula McSteen [email protected] or Dave Jackson [email protected]. Attendance is limited and registration is required. The deadline to register was February 15th, and so registration is now closed. |
Paula McSteen
David Jackson |
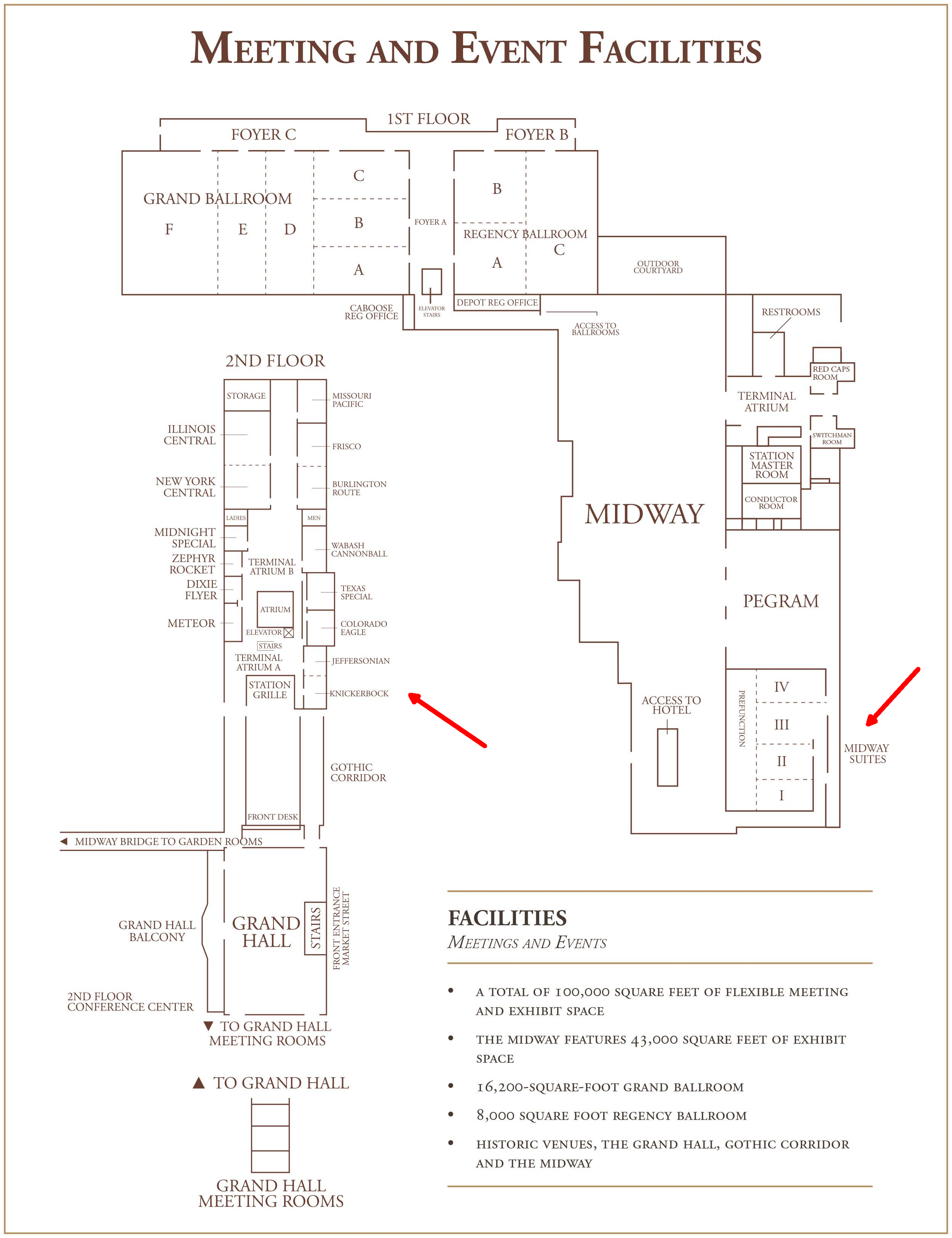
Midway 1-4 |
| 1:00 PM - 2:00 PM | MaizeMine
Join us for a tutorial on using MaizeMine, the InterMine data warehouse that now hosts B73 v5 and the 25 NAM founder genomes. |
Chris Elsik |
Jeffersonian/Knickerbock rooms |
| 2:00 PM - 3:00 PM | Gramene: Maize Pan-genome Resources
The Gramene database (http://www.gramene.org) is an integrated resource for comparative genome and functional analysis in plants. The database provides agricultural researchers and plant breeders with valuable biological information on genomes and plant pathways of numerous crops and model species - including maize - thus enabling powerful comparisons across species. During this workshop, it will be demonstrated how to query the available data at Gramene through our versatile search interface to help users search, interpret and explore public data through gene-focused views of genomic context, phylogenetic trees, gene expression profiles, pathways, and cross-referenced resources. In addition, participants will learn how to navigate the website and review the available resources at Gramene, including curated pathways and orthologous pathway projections for 120 species via the Plant Reactome, genetic variation and QTL data, and gene expression via the EBI-Expression Atlas. Besides maize, the current data release 66 of Gramene contains an additional 127 reference genomes, 6.5 million gene annotations, and 152K gene family trees, which permit comparative analyses such as sequence and syntenic comparisons, including the B73 reference compared to sorghum, rice, Brachypodium, and wheat. In recent years, we added EMS-induced mutations in sorghum and wheat. Also, manually curated maize V5 gene models. In recent years, Gramene has positioned itself as a pan-genome resource dedicated to the study of individual crop groups (e.g., maize, rice, sorghum, and grape). The Gramene Maize pan-genome site (https://maize-pangenome.gramene.org) hosts a total of 39 maize genomes including 3 assembly versions of B73, 25 NAM founders, Ab10, and 4 European genomes, plus 7 outgroups, and maize-centric gene trees, in addition to gene expression profiles and pathway associations for B73. This data as well as the contents of the FTP site will be reviewed during the session. Gramene is supported by USDA-ARS (1907-21000-030-00D) award.
|
Nicholas (Nick) Gladman
Zhenyuan (Jerry) Lu |
Jeffersonian/Knickerbock rooms |
| 2:00 PM - 4:30 PM | Danforth Center Tour
Come visit the Donald Danforth Plant Science Center, the largest non-profit plant science research institute in the world! This trip includes a detailed tour of the Danforth Center and a career panel with Danforth Center researchers and nearby 39N Innovation District members. Priority for early-career researchers (undergraduate, graduate, post-docs, and early-career faculty and industry researchers). Space is limited to first 20 people who register here. Transportation included.
|
Katie Murphy |
Meet at the Union Station front desk, and we'll bus over at 2:00pm |
| 3:15 PM - 3:45 PM | MaizeGDB: Protein Structure Resources
This worksop presents new tools for protein search and comparison based on structural superpositions. These tools leverage recent technological breakthroughs in protein structure prediction, such as the release of AlphaFold and ESMFold, which have reduced the structural biology bottleneck by several orders of magnitude. The protein search tool allows for the identification of homologous proteins based on structural superpositions, while the comparison tool facilitates the assessment of structural similarities and differences based on three structure and sequence-based alignment methods. Additionally, new genome browser tracks have been implemented, featuring color-coded exons based on average confidence score, providing researchers with a quick and easy way to assess gene model annotation quality. These tools are designed to assist maize researchers in assessing functional homology and gene annotation, as well as other information unavailable to maize scientists even a few years ago. We propose to use these tools in combination with a new method called Functional Annotations using Sequence and Structure Orthology (FASSO) to obtain a more accurate and complete set of orthologs across diverse species. We will demonstrate the use of these tools and datasets at the workshop, and show how they can be applied to search, compare, and annotate proteins between five plant species (maize, sorghum, rice, soybean, Arabidopsis) and three distance outgroups (human, budding yeast, and fission yeast). |
Carson Andorf |
Jeffersonian/Knickerbock rooms |
| 3:45 PM - 4:45 PM | MaizeGDB: Pan-genome Resources and Visualization
This workshop will provide an update on the latest genomic data available at MaizeGDB, including the NAM, PanAnd (first round), and CAAS projects. We will first define key terms such as pan-genome, pan-gene, and core set. We will then introduce the project pages for each of these datasets and the tabs for accessing metadata, browsers, and other tools. Additionally, we will cover the construction of lifted gene models across maize JBrowse browsers and how to use the tracks. The pipeline used to lift gene models will be described and demonstrated. The construction of pan-gene sets for gene model pages will also be discussed. We will provide an overview of Pandagma, a new pipeline for generating pan-gene sets, and discuss differences from lifted over browser data. Applications and uses of the data will also be discussed. Finally, information on data availability at MaizeGDB will be provided, including how to download data and explore pan-gene data, as well as an introduction to a new browsing and visualization tool available at MaizeGDB called the Genome Context Viewer (GCV). |
Maggie Woodhouse
Ethy Cannon |
Jeffersonian/Knickerbock rooms |
Location:
The Maize Development Genetics workshop will take place in the Union Station Hotel on the main floor in Midway 1-4, and the MaizeGDB workshop will take place in the Jeffersonian and Knickerbock rooms. Click the image below to see a map of the hotel conference area.
Map
|
|
|
|