Pre-meeting Events on Wednesday February 28th, and Thursday February 29th!
This year, six pre-conference workshops are being offered prior to the start of the meeting! Registration is required for the Genetics of Maize-Microbe Interactions, Development & Cell Biology, and Corn Breeding Reserach workshops.
2024 Schedule:
| Wednesday, February 28, 2024 |
Contact: |
Location: |
| 8:00 AM - 12:00 PM | Maize Crop Germplasm Committee
The Maize Crop Germplasm Committees (CGC) serve as subject matter experts to guide NPGS curatorial staff on best practices, including the priorities and techniques for characterizing the collections. They also help review proposals that fund plant explorations and evaluation grants for scientific rigor. Contact Bill Tracy for more information. |
Bill Tracy |
Room 302C |
| 1:00 PM - 6:00 PM | Corn Breeding Research.
The annual Corn Breeding Research meeting will take place Feb 28th - 29th. Registration is required. Schedule and speakers are being finalized; for more information please contact Jim Holland and Joe Gage. |
Jim Holland
Joe Gage |
Room 302A/B |
| 7:00 PM - 10:00 PM | Genetics of Maize-Microbe Interactions
A one-day Genetics of Maize-Microbe interactions Workshop will be held immediately preceding the Maize Genetics Meeting at the Raleigh Convention Center. A social event will be held on Wednesday Feb 28 from 7-10 PM, and the workshop will be held on Thursday Feb 29 from 8:30 AM - 5:30 PM. To register for the meeting please fill in the form here. For more information please contact Peter Balint-Kurti. |
Peter Balint-Kurti |
Room 301A |
| Thursday, February 29, 2024 |
Contact: |
Location: |
| 8:00 AM - 5:00 PM | Corn Breeding Research.
The annual Corn Breeding Research meeting will take place Feb 28th - 29th. Registration is required. Schedule and speakers are being finalized; for more information please contact Jim Holland and Joe Gage. |
Jim Holland
Joe Gage |
Room 302A/B |
| 8:30 AM - 5:30 PM | Genetics of Maize-Microbe Interactions
A one-day Genetics of Maize-Microbe interactions Workshop will be held immediately preceding the Maize Genetics Meeting at the Raleigh Convention Center. A social event will be held on Wednesday Feb 28 from 7-10 PM, and the workshop will be held on Thursday Feb 29 from 8:30 AM - 5:30 PM. To register for the meeting please fill in the form here. For more information please contact Peter Balint-Kurti. |
Peter Balint-Kurti |
Room 301A |
| 9:00 AM - 4:00 PM | Development and Cell Biology
Join us for a workshop dedicated to discussing the latest research in Maize Development and Cell Biology. Graduate students, postdocs and junior faculty will present their results in a short talk format, with a small, friendly audience. Plenty of time will be available after each talk for animated discussion. If you would like to present your work, or if you have any questions, please email the workshop organizers. Attendance is limited and registration is now closed. Please do not sign up unless you will be able to attend the entire workshop from 9am-4pm on Thursday Feb 29th. Deadline to register is Monday January 29, 2024.
|
Paula McSteen
David Jackson
Josh Strable |
Room 301B |
| 3:00 PM - 4:00 PM | Gramene Workshop
The Gramene database is an integrated resource for comparative genome and functional analysis in plants. The database provides agricultural researchers and plant breeders with valuable biological information on genomes and plant pathways of numerous crops and model species - including maize - thus enabling powerful comparisons across species. During this workshop, it will be demonstrated how to query the available data at Gramene through our versatile search interface to help users search, interpret and explore public data through gene-focused views of genomic context, phylogenetic trees, gene expression profiles, pathways, and cross-referenced resources. Molecular biologists, geneticists, and genomicists wiIl find value in the expansive phylogenetic trees and incorporated homology views and information for genes and gene families of interest. These features are also integrated with greater-detailed web pages that host easy-to-access sequence information, ontologies, and a genome browser application that contain permanent expression and variation information as well as the ability to load custom browser tracks to assist in users' genomic experimentation. One of the more powerful features of Gramene and its pan-genome subsites is the curation of natural and EMS-induced variant population data within certain species; users can query gene models for disruptive variants and identity which respective population and individual germplasm contain these single nucleotide polymorphisms or structural variants. Additionally, the search interface allows geneticists and breeders to query a broad swath of public data for available gene models within species, including QTLs, ontologies, and metabolic pathway affiliations. In recent years, Gramene has positioned itself as a pan-genome resource dedicated to the study of individual crop groups (e.g., maize, rice, sorghum, and grape). The Gramene Maize pan-genome site hosts a total of 39 maize genomes including 3 assembly versions of B73, 25 NAM founders, Ab10, and 4 European genomes, plus 7 outgroups, and maize-centric gene trees, in addition to gene expression profiles and pathway associations for B73. All of these data are available for download via a FTP site and can be queried in bulk with multiple filters through the GrameneMart portal. Gramene is supported by USDA-ARS (1907-21000-030-00D) award.
|
Doreen Ware
Nick Gladman
|
Room 304 |
| 4:00 PM - 5:30 PM | MaizeGDB: Pan-genome and AI resources
Join us for a workshop dedicated to the new pan-genome and AI resources at MaizeGDB! The first half of the workshop will focus on new pan-genome updates at MaizeGDB. Our update includes comprehensive features like the PanAnd genomes showcasing 13 Andropogoneae tribe grasses, extensive pan-genes information, a dedicated gene center webpage, and other new features. The first-round release of the PanAnd Consortium project is now available, offering detailed genomes and annotations. We've also performed an in-depth Zea pan-gene analysis on 58 maize genomes, with each analysis's details accessible through specialized tabs, enriched with various visual tools for exploring pan-genes. Our gene center provides robust functionalities for gene locus and model searches, bulk downloads, and a wealth of gene-related data. The second half of the workshop will describe new artificial intelligence and machine learning resources found at our AI data center. This data center is a hub for various AI tools, methodologies, and datasets, including functional predictions, protein embeddings, and new tools for protein structure comparison and mutation impact analysis across the maize pan-genome.
|
Carson Andorf
|
Room 304 |
Location:
All workshops will take place on the main level (Rooms 300+) of the Raleigh Convention Center. Click the image below to see a map of the conference area.
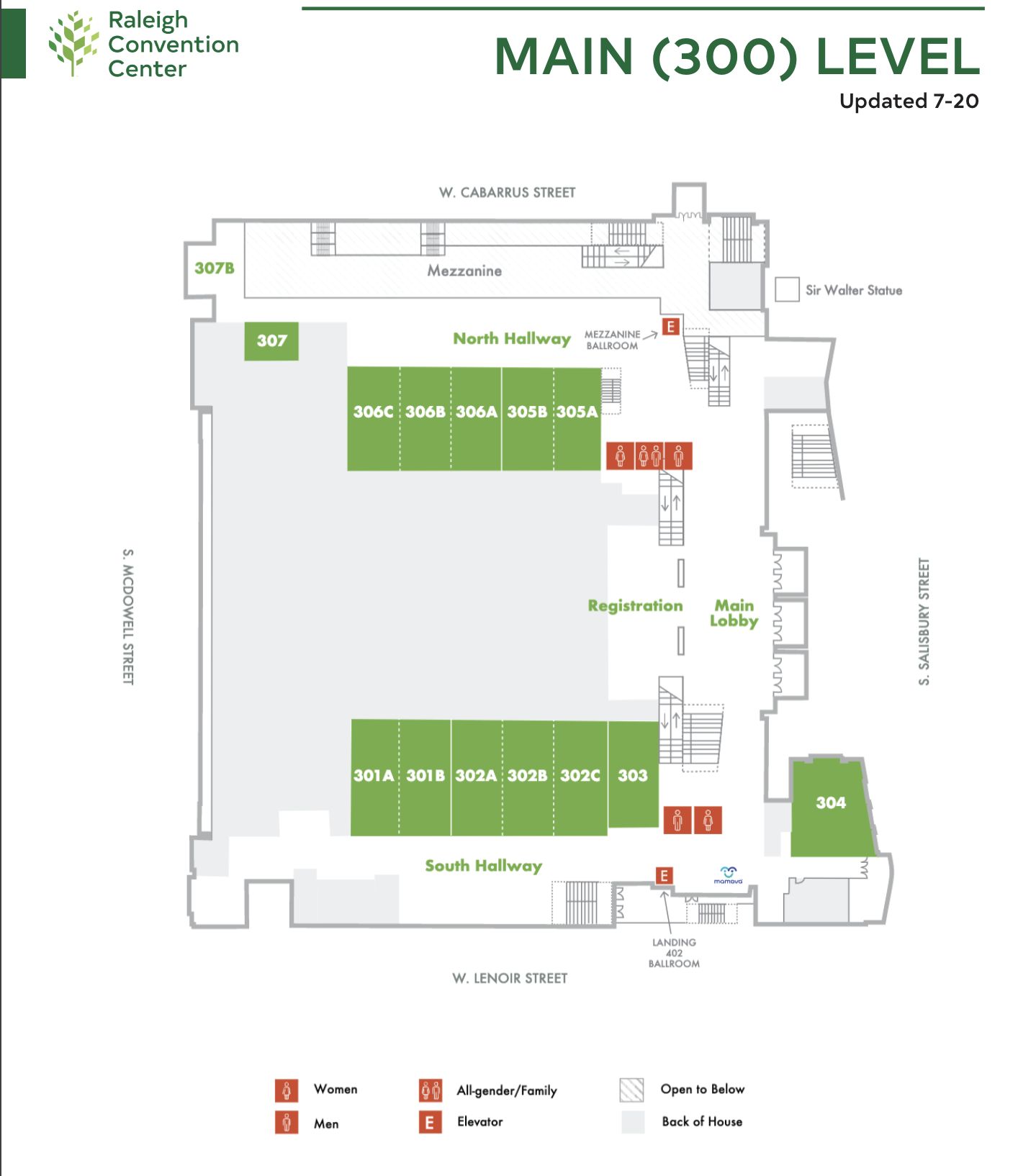
Map
|
|
|
|